Accessing PDTDB
Searching PDTDB
- Keyword (or) Accession Number Search
- 2D Chemical Structure Similarity Search
- Similar Chemical Structure Search by Formula
- Similar Binding Search
Browsing PDTDB
Downloading from PDTDB
Structure Visualization
Properties Prediction
Keyword (or) accession number search
The textbox in the PDTDB search page is enabled with AJAX features. So, just by typing the letters we can able to easily autocomplete the keyword or accession number in the textbox.
Go to PDTDB search page. To search database by keywords (or) accession number, click the “Keyword Search” (or) “Unique ID Search” tab and select “Target Details” (OR) “Plant Details” (OR) “Ligand Details” (OR) “Docking Details”. Enter the keyword in the textbox and click the “SEARCH” button.
2D Chemical Structure Similarity Search
“2D Chemical Structure Similarity Search” method is similar to “Keyword (or) accession number search” method. Additionally, you can able to search chemical structure in PDTDB or PubChem database. To search chemical structures in the database, you must draw the chemical structure in the JSDraw (a Java chemical structure editor) which will automatically input SMILES string in the textbox.
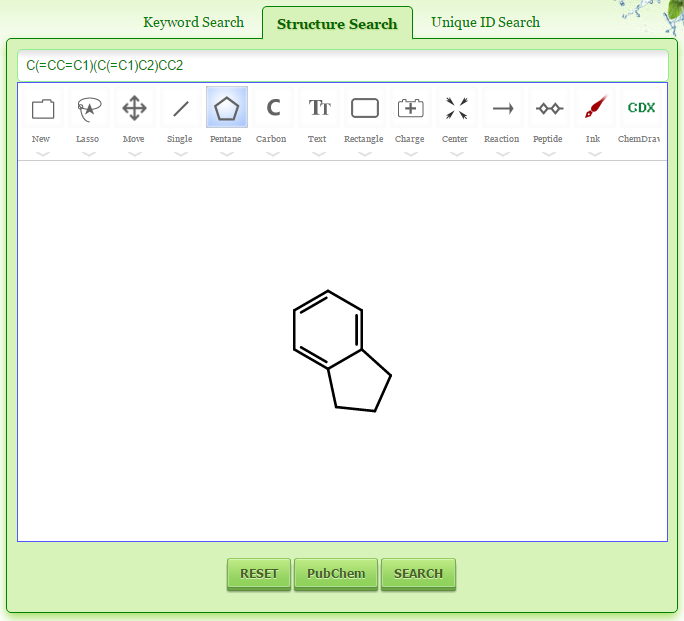
Go to PDTDB search page. To search the database by SMILES string, click the “Structure Search” tab and enter the SMILES string in the textbox and click the “SEARCH” button.
Go to PDTDB search page. To search the database by drawing chemical structure, click the “Structure Search” tab and then click the “JSDraw” button. Draw the chemical structure in the JSDraw tool and click the “SEARCH” button. While drawing the chemical structure, the SMILES string is generated instantly in the textbox. Similarly click the “PubChem” button if you want to search the PubChem database.
Similar Chemical Structure Search by Formula
Each output page of the ligand database / docking database has a “Molecular Formula” entity with hyperlinked chemical structure formula string. Just by clicking the hyperlink, you can able to search chemical structures having similar chemical structures.
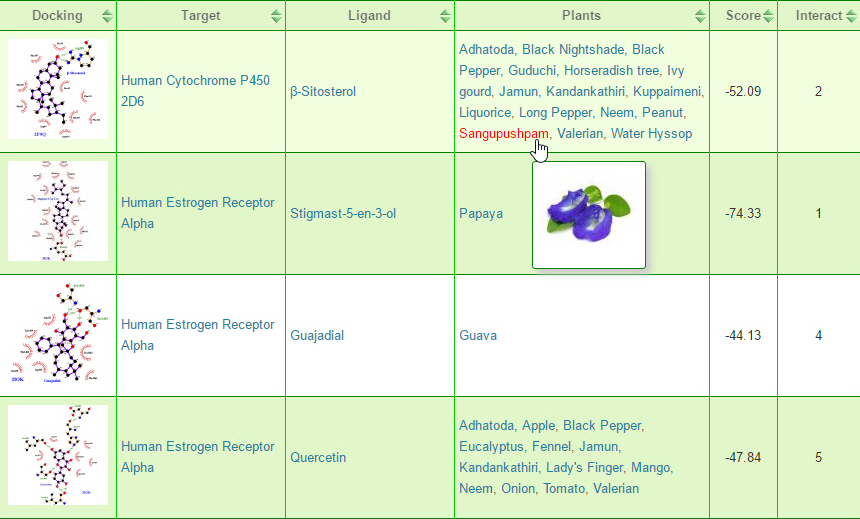
In PDTDB, the link to binding page is located in the search menu in the menu bar. By moving the cursor over the search menu expands and the submenu will be visible. Binding search page allows users to search for drug target and ligand complex structures using target or ligand keyword. The search result provides target-ligand interaction plot, a list of matching targets or ligands based on your query, and list of plants containing the ligands.
Binding search allows users to predict the structure activity relationship (SAR) of the biomolecules. Currently PDTDB supports target-wise relationship and ligand-wise relationship prediction of biomolecules. The result page of binding search is displayed as a sortable table containing target-ligand interaction, target details, ligand detail, a list of plants containing the ligand, docking score, and target-ligand interaction bonds.
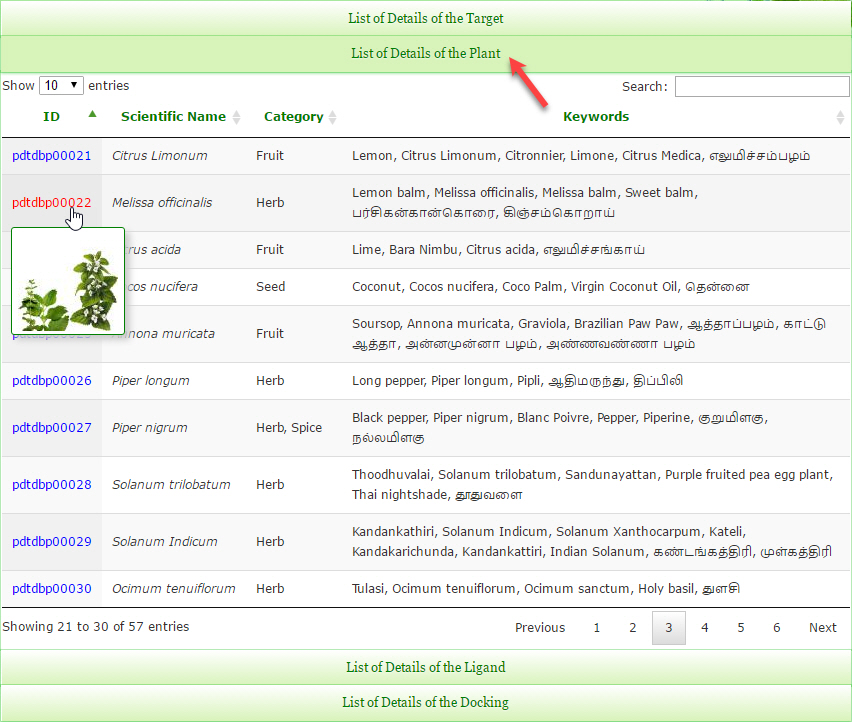
In PDTDB browse page, the list of databases is displayed in the accordion option format. So, just by clicking the accordion header the records in the particular database is highlighted and remaining database are hidden.
In PDTDB browse page, the records of each database are displayed in a sortable table format. So, just by clicking the table column header the records are sorted in ascending or descending order accordingly.
“Browse Database by Filter” method is similar to the “Browse Database by Sort” method. Additionally, the large number of rows in the table can be filtered by typing the keyword in the textbox.
In each output page of the target database / ligand database / docking database there are two buttons “View in 3D” or “Download”. By clicking the “Download” button, structure is downloaded automatically.
In each output page of the target database / docking database there is a download icon near the “Target Sequence :” entry. By clicking the left icon, the target sequence in the fasta file format is downloaded automatically.
The PDTDB download page allows you to download the sequences and structures as batches in compressed file format (ZIP). In batch download, the textbox is enabled with AJAX features. So, just by typing the letters we can able to easily autocomplete the accession number in the textbox.
Go to PDTDB download page. To download the target or docking structure, click the “STRUCTURES” tab and choose the option “Structure from Target Database” or “Structure from Docking Database”. Enter the target or docking database accession number in Comma Seperated Values (CSV) format in the textbox and click the “Download” button. To download the ligand structure, click the “LIGANDS” tab and enter the ligand database accession number in Comma Seperated Values (CSV) format in the textbox and click the “Download” button. Similarly, target sequences are downloaded from the “SEQUENCES” tab.
In the PDTDB download page, the summary of target database, plant database, ligand database, and docking database entries can be downloaded in CSV file format, which is compatible to view using Excel of Microsoft Office.
Go to PDTDB download page. Click the “SUMMARY” tab and click the hyperlink “here” in the list of the lines based on the choice of the database.
Protein (or) Ligand Visualization
In each output page of the target database there are two buttons “View in 3D” or “Download” in right side as shown in “Direct Download” section. By clicking the “View in 3D” button, the target structure is displayed in JSDraw (a Java molecule visualization tool) in cartoon model.
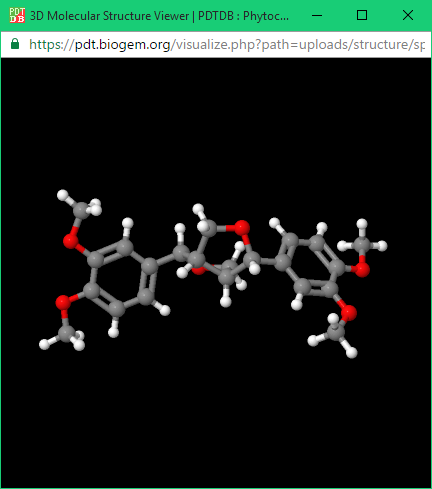
In each output page of the ligand database there are two buttons “View in 3D” or “Download” in right side as shown in “Direct Download” section. By clicking the “View in 3D” button, the ligand structure is displayed in JSDraw (a Java molecule visualization tool) in ball and stick model.
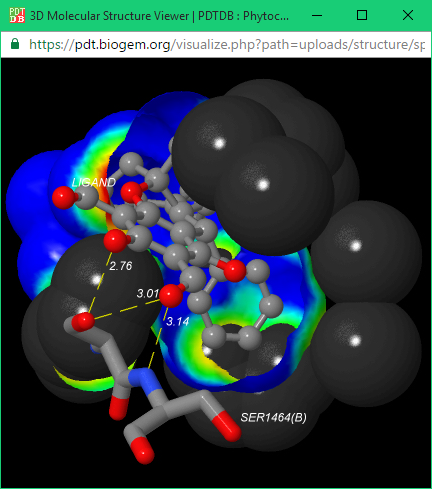
Protein-Ligand Interaction Visualization
In each output page of the docking database there are two buttons “View in 3D” or “Download” in right side as shown in “Direct Download” section. By clicking the “View in 3D” button, the docking structure is displayed in JSDraw (a Java molecule visualization tool) in target-ligand interaction model.
In each output page of the target database / docking database there is a information icon near the “Target Sequence :” entry. Towards right, the first icon is for downloading target sequence in the fasta file format, second icon for predicting physicochemical properties of the target sequence, and third icon for predicting related 3D protein structures from the Protein Data Bank. By clicking the second icon, the physicochemical properties such as length, molecular weight, theoretical isoelectric point (pI), amino acid composition, total number of negatively and positively charged amino acids, atomic composition of amino acids, molecular formula, total number of atoms in the protein sequence, extinction coefficients, estimated half-life, instability index (II), aliphatic index, aromaticity (Y+W+F), grand average of hydropathicity (GRAVY), and Kyte & Doolittle hydrophobicity plot of the protein sequence can be predicted.
A model Kyte & Doolittle hydrophobicity plot of the protein sequence (pdtdbt00003) is given below.
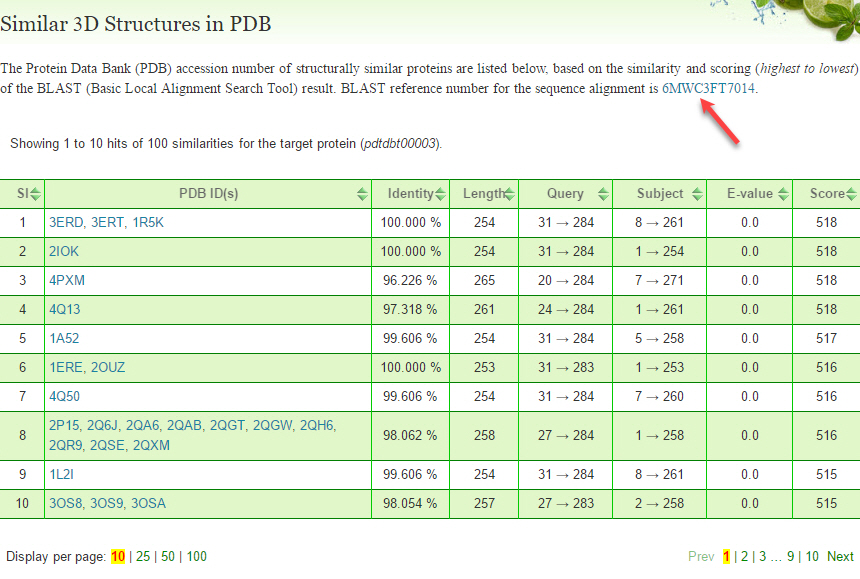
A model BLAST (Basic Local Alignment Search Tool) result for similar 3D protein structures in PDB (Protein Data Bank), matching the protein sequence (pdtdbt00003) is given below. By clicking the hyperlinked reference number, the page redirects to the detailed interactive multi-featured graphical output of the NCBI BLAST page.